Gene Ontology Reorganized, Resistance to Imatinib, and Single-Cell Expression Data: the PLOS Comp Biol November Issue
Header Image Credit: Maesani et al.
Here are our highlights from November’s PLOS Computational Biology:
Finding New Order in Biological Functions from the Network Structure of Gene Annotations
The Gene Ontology (GO) provides biologists with a controlled terminology that describes how genes are associated with functions and how functional terms are related to one another. These term-term relationships encode how scientists conceive the organization of biological functions, and they take the form of a directed acyclic graph (DAG). Kimberly Glass and Michelle Girvan propose that the network structure of gene-term annotations made using GO can be employed to establish an alternative approach for grouping functional terms that captures intrinsic functional relationships that are not evident in the hierarchical structure established in the GO DAG.
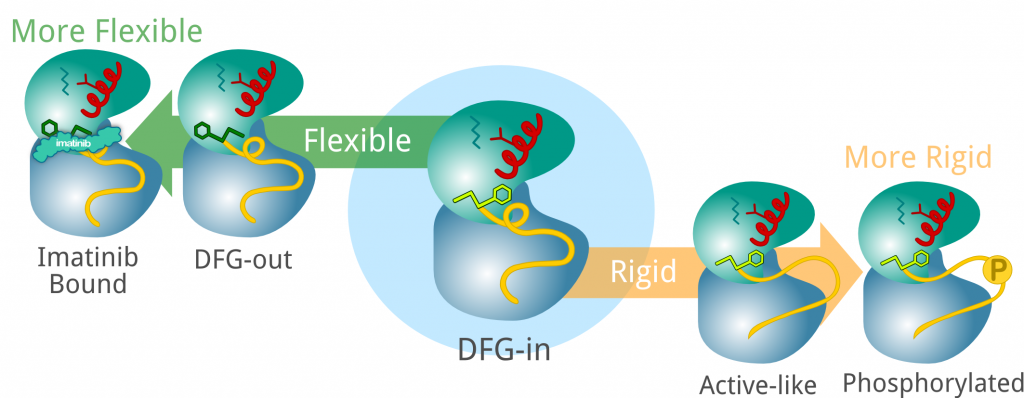
Towards a Molecular Understanding of the Link between Imatinib Resistance and Kinase Conformational Dynamics
Imatinib remains the most important and studied anti-cancer drug for cancer therapy in its new paradigm. Due to its inhibition of the Abl kinase domain, imatinib is strikingly effective in the initial stage of chronic myeloid leukemia with more than 90% of the patients showing a complete remission. However, the emergence of drug resistance is a serious concern. Francesco L. Gervasio and colleagues investigate the molecular mechanism of drug-resistant mutations which, despite the importance and the adverse effect on cancer patients’ prognosis, is still debated.
SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis
A major challenge in developmental biology is to understand the genetic and cellular processes driving organ formation and differentiation of the diverse cell types that comprise the embryo. Yan Xu and colleagues present SINCERA, a generally applicable analytic pipeline for processing scRNA-seq data from a whole organ or sorted cells. The pipeline supports the analysis for: 1) the distinction and identification of major cell types; 2) the identification of cell type specific gene signatures; and 3) the determination of driving forces of given cell types. SINCERA is freely available from https://research.cchmc.org/pbge/sincera.html.