Modelling Atherosclerotic Plaque Formation, Tracking Neurons, and Timestamping Molecular Recorders: the PLOS Comp Biol May issue
Check out our highlights from the PLOS Computational Biology May issue:
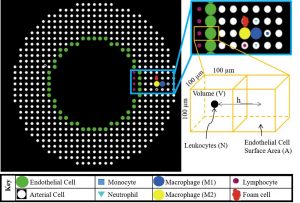
An agent-based model of leukocyte transendothelial migration during atherogenesis
Atherosclerosis affects millions of people worldwide and is characterized by a maladaptive build-up of fatty material, leukocytes, and extracellular matrix inside the artery wall. With age this material, collectively called a plaque, enhances and blocks blood flow thereby altering the hemodynamics. If the plaque ruptures, the occlusion may cause a life-threating stroke or myocardial infarction. Rita Bhui and colleagues developed an agent-based model capable of integrating cell behaviors and hemodynamic properties as the plaque grows, and identified crucial lumen volumes that give rise to favorable conditions for rapid plaque growth. Collectively, understanding how mechanobiological events are integrated within an artery will help elucidate emergent behaviors and predict plaque evolution.
Automatically tracking neurons in a moving and deforming brain
Computer algorithms for identifying and tracking neurons in images of a brain have struggled to keep pace with rapid advances in neuroimaging. In small transparent organisms like the nematode Caenorhabditis elegans, it is now possible to record neural activity from all of the neurons in the animal’s head with single-cell resolution as it crawls. A critical challenge is to identify and track each individual neuron as the brain moves and bends. In this work, Andrew M. Leifer and colleagues present a fully automated algorithm for neuron segmentation and tracking in freely behaving C. elegans. The approach uses non-rigid point-set registration to construct feature vectors that describe the location of each neuron relative to other neurons and other volumes in the recording, and then clusters feature vectors in a time-independent fashion to track neurons through time.

Nucleotide-time alignment for molecular recorders
This work presents a computational tool needed for the development and implementation of molecular recorders, a promising potential technique for massive-scale neuroscience. Molecular recorders use proteins to encode levels of a substance of interest as detectable changes in a linear cellular structure, much like a ticker tape represents information linearly on a strip of paper. However, molecular recorders suffer a particular drawback involving timing: unlike most methods of recording signals, in simple molecular recording systems one does not observe when each data point was recorded. This timing information is almost always required in order to make associations between the recorded data and the rest of the experiment. In this work, Thaddeus R. Cybulski and colleagues propose a method to estimate the timing of these data points using easily-observable experimental measurements. The authors demonstrate the application of this method in a simulated neuroscience paradigm, investigate the effects of experimental design, and determine protein properties that would be desirable in molecular recorders. These findings are useful both as a computational proof-of-concept, and as guidelines for current efforts to engineer proteins for molecular recording.
Header Image Credit: Nguyen et al.
