Inferring Gene Regulatory Networks; Designing Antibiotic Cocktails and Modelling Synaptic Depression
Check out our Editors-in-Chief’s selection of papers from the January issue of PLOS Computational Biology. The first one of 2019!
Multi-study inference of regulatory networks for more accurate models of gene regulation
Due to an increasing availability of biological data, methods to properly integrate data generated across the globe become essential for extracting reproducible insights into relevant research questions. In this work, Castro et al. developed a framework to reconstruct gene regulatory networks from expression datasets generated in separate studies—and thus, because of technical variation (different dates, handlers, laboratories, protocols etc…), challenging to integrate.
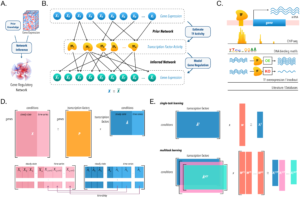
Since regulatory mechanisms are often shared across conditions, the authors hypothesized that drawing conclusions from various data sources would improve the performance of gene regulatory network inference. By transferring knowledge among regulatory models, their method is able to detect weaker patterns that are conserved across datasets, while also being able to detect dataset-unique interactions.
Prediction of ultra-high-order antibiotic combinations based on pairwise interactions
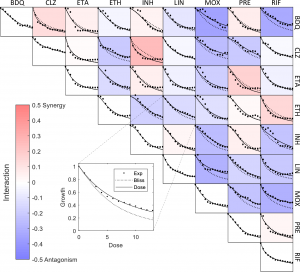
Drug combinations are important to increase efficacy and reduce the resistance of treatment for infection and cancer. The major challenge is the vast number of experiments needed to scan the space of combination in order to find rare synergistic drugs and their optimal doses. In the past few years, there has been an advance in the ability to predict the effects of drug cocktails, using a small number of experiments on drug pairs. These approaches have not been tested on combinations of more than a few drugs. Thus, it remains unclear whether there are useful combinations of 5–10 drugs that work at low doses. Here Katzier et al. show that a mathematical model can use data for drug pairs to predict ultra-high-order cocktails for E. coli and an important pathogen, M. tuberculosis.
Short-term synaptic depression can increase the rate of information transfer at a release site
Fatigue is an intrinsic property of living systems and synapses are no exception. Synaptic depression reduces the ability of synapses to release vesicles in response to an incoming action potential. Whether synaptic depression simply reflects the exhaustion of neuronal resources or whether it serves some additional function is still an open question. Salmasi et al. ask how synaptic depression modulates the information transfer between neurons by keeping the synapse in an appropriate operating range. Using a tractable mathematical model for synaptic depression of both synchronous spike-evoked and asynchronous release of neurotransmitter, they derive a closed-form expression for the mutual information rate. Depression, it turns out, can both enhance or impair information transfer, depending on the relative level of depression for synchronous spike-evoked and asynchronous releases.
