This week in PLOS Biology
In PLOS Biology this week you can read about the perception and self-regulation of pain, a computational model of operant learning and improving the computer-friendliness of phenotypic data.
Two Parallel Pathways to Pain
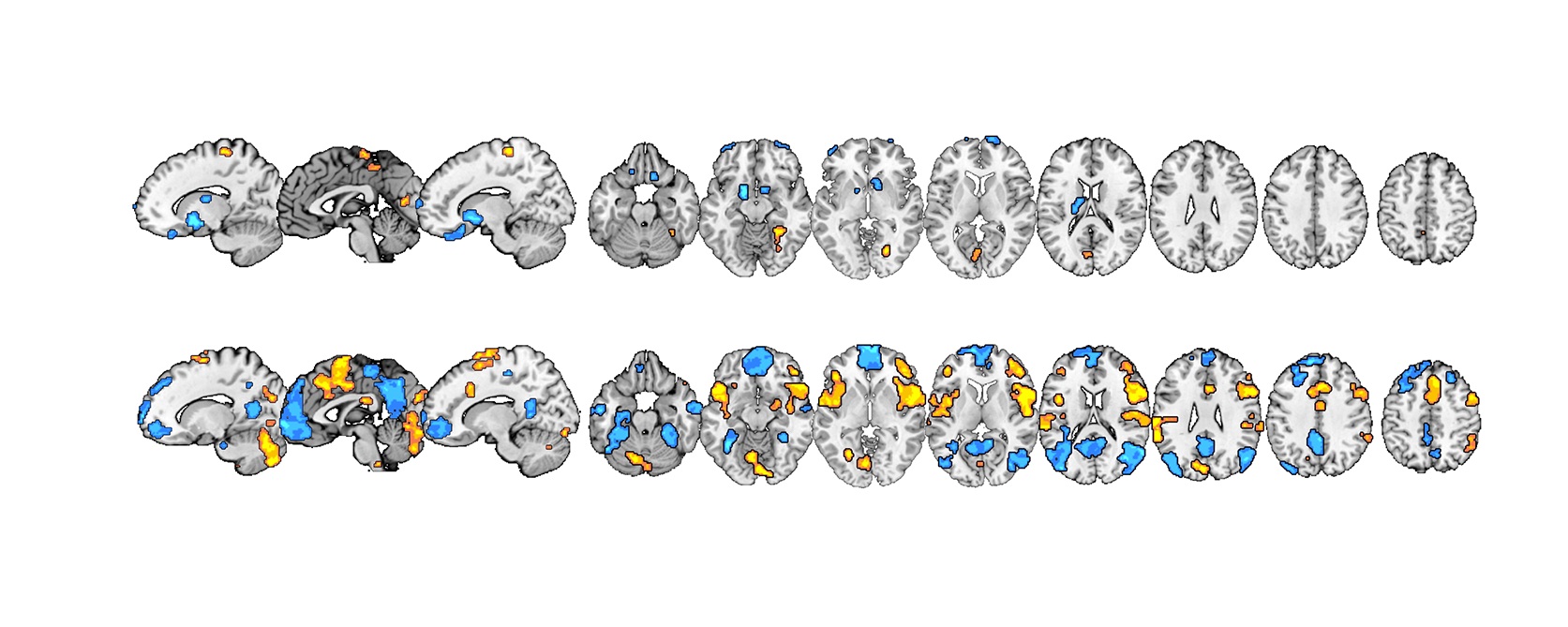
Image credit: 10.1371/journal.pbio.1002036
Understanding how pain is processed in the brain is a controversial area of neuroscience, because there doesn’t seem to be a single ‘‘pain cortex’’ that directly codes the subjective perception of pain. Although we’ve known for some time that we can self-regulate pain we experience, it is unclear which circuits underpin this. Choong-Wan Woo, Tor Wager & colleagues used fMRI to show that two distinct parallel neural systems independently contribute to our overall experience of pain — separately modulated by noxious input and by cognitive self-regulation. Read more in the accompanying Primer.
How Animals Select Actions with Rewarding Outcomes
A key component of survival is learning to associate rewarding outcomes with specific actions, such as searching for food. Actions are represented in the cortex, and rewarding outcomes activate neurons that release dopamine. These signals are then sent to the striatum— the input station for a collection of brain structures called the basal ganglia, which play an important role in action selection. A new paper by Kevin Gurney, Mark Humphries & Peter Redgrave uses a computational model that shows how the brain’s internal signal for outcome changes the strength of neuronal connections, leading to the selection of rewarded actions and the suppression of unrewarded ones. Read more in the accompanying Synopsis.
Computing Phenotype

doi:10.1371/journal.
pbio.1002033.g001
Phenotypic data (i.e. observable traits such as anatomy and behaviour) represent much of what we know about life and drive much of life science research. A new Perspective by Andrew Deans & colleagues argues that the current form in which phenotypic data are recorded inhibits their productive use. Their article asks us to imagine a future in which we could compute across phenotype data as easily as genomic data, calls for efforts to realize this vision, and discusses the potential benefits.
