Brain Network Eigenmodes in Health and Disease, Olfactory Learning Without Mushroom Bodies, and Assessing Overlap of Genomic Features: the PLOS Comp Biol June Issue
Check out our highlights from the PLOS Computational Biology June issue:
Brain Network Eigenmodes Provide a Robust and Compact Representation of the Structural Connectome in Health and Disease
While the structural connectome of the brain has emerged as a powerful tool towards understanding the progression of neurologic and psychiatric disorders, links between the anatomy of connections within the brain and the effects of localized white matter pathology on cognition are still an active area of investigation. Here, Ashish Raj and colleagues propose the use of the diffusion process towards understanding perturbations of brain connectivity. The authors find that while the dynamics of this process are strongly conserved in healthy subjects, they display significant and interpretable deviations in individuals with agenesis of the corpus callosum — one of the most common brain malformations. These findings, including the strong similarity between regions identified to be crucial for diffusion and nexus regions of white matter from edge density imaging, show converging evidence towards understanding the relationship between white matter anatomy and the structural connectome.
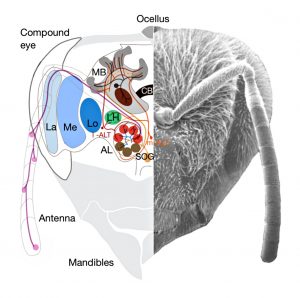
Olfactory Learning Without the Mushroom Bodies: Spiking Neural Network Models of the Honeybee Lateral Antennal Lobe Tract Reveal its Capacities in Odour Memory Tasks of Varied Complexities
The honeybee olfactory system offers the opportunity to study different levels of learning complexity in a small network. Odour information is transferred from the antennae to the antennal lobes, and from there to the mushroom bodies and the lateral horn via parallel medial and lateral tracts of projection neurons. Although much progress has been made in understanding olfactory coding in the bee brain, the precise contribution of the lateral pathway in olfactory learning is still unclear. To understand the computational mechanisms underpinning the lateral antennal lobe tract, Lars Chittka and colleagues modelled local computation and non-associative plasticity within glomeruli in the antennal lobe and the lateral horn where they were additionally modulated by the neurotransmitter octopamine to achieve associative learning. They establish that the connectivity within the antennal lobe (shaped by non-associative plasticity) can also be modified by inhibitory feedback neurons, making output patterns of the antennal lobe more separable and sparser. The authors’ modelling indicates that bees, using the lateral pathway, might learn to solve positive patterning tasks in addition to elemental learning and olfactory generalisation without the contribution of the mushroom bodies. The model can’t account for negative patterning, however, indicating that the mushroom bodies may be required for this discrimination. The modelling approach allows changes in structural organization of honeybees’ antennal lobes to be linked with behavioural performance over the course of their life.
GINOM: A Statistical Framework for Assessing Interval Overlap of Multiple Genomic Features
The age of genomics has made a large number of datasets available for the wider scientific community. Many of these datasets come in the form of genomics tracks, represented as features associated with a collection of genomic intervals along chromosomes. A common tack in genomics is to identify putative associations between these features that can lead to new insights about genome organization and function. For example, the activity of certain classes of genes might be influenced by the presence of specific combinations of chromatin modifications and binding of transcription factors at their promoters or enhancers. Here, Nicola Neretti and colleagues present a novel methodology, named GINOM (Genomic INterval Overlap Model), to test for the significance of these associations. The authors apply it to the problem of detecting biases in the locations along chromosomes where mobile genetic elements tend to insert themselves, and identify a potential preference for L1 elements towards gene bodies marked by a facultative heterochromatic mark.
Header image credit: Wang et al.
